Bacteria incorporate pieces of old DNA in their own genome, scientists discover
What DNA fragments --- some thousands of years old --- from
biological waste and wastewater have hospital bacteria incorporated?
November 19, 2013
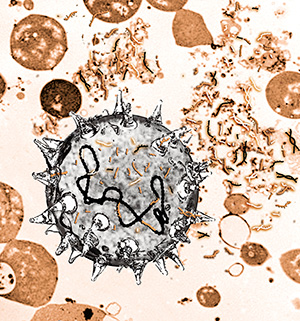
Illustration
of bacteria that take up DNA fragments from their surroundings. Some of
the grasped DNA fragments attach to the bacteria’s own DNA and are
incorporated. (Credit: Katrine Harving Holm).
This discovery may have major consequences both in connection with resistance to antibiotics in hospitals and in our perception of the evolution of life itself.
Our surroundings contain large amounts of strongly fragmented and damaged DNA, which is being degraded. Some of it may be thousands of years old.
Laboratory experiments with microbes and various kinds of DNA have shown that bacteria take up very short and damaged DNA from the environment and passively integrate it in their own genome.
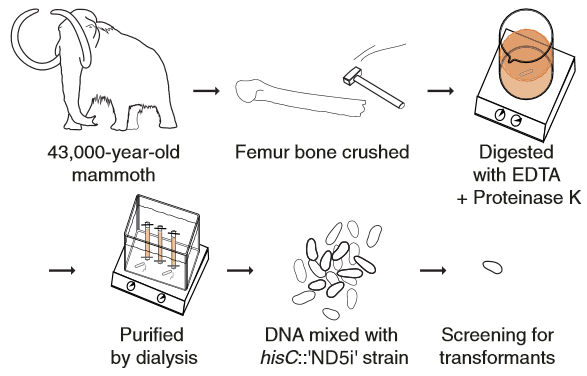
Furthermore, this mechanism has also been shown to work with a modern bacteria’s uptake of 43.000 years old mammoth DNA.
The results were just published in Proceedings of the National Academy of Sciences (open access).
Second-hand Evolution
[+]
The discovery of this second-hand use of old or fragmented DNA may have major future consequences. Postdoc Søren Overballe-Petersen from the Center for GeoGenetics at the Natural History Museum of Denmark
first author on the paper, says it is well-known that bacteria can take
up long intact pieces of DNA. “But so far the assumption has been that
short DNA fragments were biologically inactive. Now we have shown that
this assumption was wrong. As long as you have just a tiny amount of DNA
left over there is a possibility that bacteria can re-use the DNA.”
Diagram
of the ancient DNA experiment. Woolly mammoth DNA was used as donor DNA
for natural transformation of the hisC::′ND5i′ strain (credit: Søren
Overballe-Petersen et al./PNAS)
“One consequence of this is in hospitals that have persistent problems with antibiotic resistance,” says Kaare M. Nielsen from University of Tromsø in Norway. “In some cases, they will have to start considering how to eliminate DNA remnants. So far, focus has been on killing living pathogen bacteria, but this is no longer enough in the cases where other bacteria afterwards can use the DNA fragments which contain the antibiotic resistance.
“The research group’s results reveal that the large reservoir of fragments and damaged DNA in the surroundings preserve the potential to change the bacteria’s genomes even after thousands of years. This is the first time a process has been described which allows cells to acquire genetic sequences from a long gone past. We call this phenomenon “Anachronistic Evolution” or “Second-hand Evolution.”
“DNA from dead organisms drives the evolution of living cells, in contradiction with common belief of what drives the evolution of life itself,” said Professor Eske Willerslev from the Center for GeoGenetics at the Natural History Museum of Denmark, leader of the project.
How damaged DNA creates new sequences
“Furthermore, old DNA is not limited to only returning microbes to earlier states. Damaged DNA [20 or more base pairs] can also create new combinations of already functional sequences. You can compare it to a bunch of bacteria which poke around a trash pile looking for fragments they can use. Occasionally they hit some ‘second-hand gold,’ which they can use right away.
“At other times they run the risk of cutting themselves up. It goes both ways. This discovery has a number of consequences partially because there is potential risk for people when pathogen bacteria or multi-resistant bacteria exchange small fragments of ‘dangerous’ DNA, e.g., at hospitals, in biological waste and in wastewater.
“In the grand perspective the bacteria’s uptake of short DNA represents a fundamental evolutionary process that only needs a growing cell consuming DNA pieces. A process that possibly is a kind of original type of gene-transfer or DNA-sharing between bacteria. The results show how genetic evolution can happen in jerks in small units. The meaning of this is great for our understanding of how microorganisms have exchanged genes through the history of life. The new results also support the theories about gene-transfer as a decisive factor in life’s early evolution.”
“This is one of the most exciting perspectives of our discovery,” says Overballe-Petersen. “Computer simulations have shown that even early bacteria on Earth had the ability to share DNA — but it was hard to see how it could happen. Now we suggest how the first bacteria exchanged DNA. It is not even a mechanism developed to this specific purpose but rather as a common process, which is a consequence of living and dying.”
Abstract of Proceedings of the National Academy of Sciences paper
DNA molecules are continuously released through decomposition of organic matter and are ubiquitous in most environments. Such DNA becomes fragmented and damaged (often <100 43="" a="" abasic="" acinetobacter="" acquired="" additionally="" adjacent="" among="" ancient="" and="" appear="" are="" as="" bacteria.="" bacteria="" bacterial="" bacterium="" baylyi="" be="" bone="" bp="" but="" by="" can="" common="" contain="" cross-links="" damaged="" demonstrate="" directly="" dna="" double-nucleotide="" environment="" environmental="" escape="" events="" evolution.="" exchange="" find="" findings="" for="" fragmented="" fragments="" from="" further="" generated="" genetic="" genomes="" half="" here="" hundreds="" in="" include="" including="" independent="" is="" large="" lesions="" linked="" mammoth="" may="" microbes="" million="" miscoding="" mismatch="" molecules.="" molecules="" more="" moreover="" natural="" nontransformable="" not="" nucleotide="" nutrient="" occur="" of="" old.="" open="" or="" our="" p="" persist="" polymorphisms="" possibility="" potential="" present="" quantities="" reca="" recognized="" recombinase="" recombination="" repair.="" replication.="" reveal="" several="" short="" show="" sites="" source="" substrate="" such="" than="" that="" the="" thousands="" through="" to="" transformable="" transformation.="" transformation="" truly="" up="" uptake="" variations="" we="" which="" with="" woolly="" y-old="" years.="" years="">
(¯`*• Global Source and/or more resources at http://goo.gl/zvSV7 │ www.Future-Observatory.blogspot.com and on LinkeIn Group's "Becoming Aware of the Futures" at http://goo.gl/8qKBbK │ @SciCzar │ Point of Contact: www.linkedin.com/in/AndresAgostini
 Washington
Washington