Radical genome recoding
First-ever entirely genomically reprogrammed organism
October 24, 2013
[+]
In two parallel projects by Harvard’s Wyss Institute, researchers have created new genomes inside the bacterium E. coli in
ways that test the limits of genetic reprogramming and open new
possibilities for increasing flexibility, productivity. and safety in
biotechnology.
Wyss Institute file photo of E. Coli (credit: Rick Groleau/Wyss Institute)
The work was led by Dr. George Church, Robert Winthrop Professor of Genetics at Harvard Medical School and founding core faculty member at the Wyss Institute for Biologically Inspired Engineering. Farren Isaacs, assistant professor of molecular, cellular, and developmental biology at Yale School of Medicine, is co-senior author on the first study.
“This is the first time that a cellular genome has been designed and built in order to change functioning of that cell,” Church explained to KurzweilAI. “In particular, we changed the genetic code previously common to essentially all of life. We did this to enable use of a variety of non-standard amino acids and simultaneously make cells resistant to viruses.
“We did this in a cell that is considered very industrially useful. We made the genome changes is a way that allows radical changes to essential genes. Previous work typically involved non-essential genes and/or minor changes. This was also a much larger genome than previous attempts (4.7 million base pairs).”
Radical genomic recoding
[+]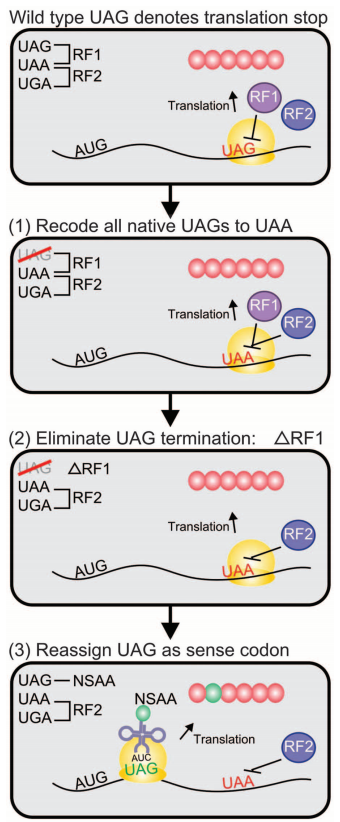
In one project, the researchers created a novel genome — the
first-ever entirely genomically recoded organism — by replacing all 321
instances of a specific “genetic three-letter word,” called a codon,
throughout the organism’s entire genome with a word of supposedly
identical meaning.
Engineering
a genomically recoded organism (GRO) with a reassigned UAG codon (the
most common start codon) (credit: Marc J. Lajoie et al./Science)
The researchers then reintroduced a reprogrammed version of the original word (with a new meaning, a new amino acid) into the bacteria, expanding the bacterium’s vocabulary and allowing it to produce proteins that do not normally occur in nature.
In the second project, the researchers removed every occurrence of 13 different codons across 42 separate E. coli genes, using a different organism for each gene, and replaced them with other codons of the same function. When they were done, 24 percent of the DNA across the 42 targeted genes had been changed, yet the proteins the genes produced remained identical to those produced by the original genes.
“The first project is saying that we can take one codon, completely remove it from the genome, then successfully reassign its function,” said Marc Lajoie, a Harvard Medical School graduate student in the lab of George Church. “For the second project we asked, ‘OK, we’ve changed this one codon, how many others can we change?’”
Of the 13 codons chosen for the project, all could be changed.
“That leaves open the possibility that we could potentially replace any or all of those 13 codons throughout the entire genome,” Lajoie said.
Important commercial applications
The research has important commercial applications, Church told KurzweilAI. “Viral infection can not only ruin huge vats of growing commercial cells, but even shut down the whole factory for many months. So a strain resistant to viruses would be more productive and safer.
“Some relevant synthetic genomics technologies are [already] available from Gen9bio.com The genome and cell computer-aided-design (CAD) tools which interface with next-generation sequencing will be available this year. The devices for reading and writing genomes are getting close to hand held size [with] commercial versions within a year or so.”
Toward safer, more productive, more versatile biotech
Recoded genomes can confer protection against viruses — which limit productivity in the biotech industry — and help prevent the spread of potentially dangerous genetically engineered traits to wild organisms.
“In science we talk a lot about the ‘what’ and the ‘how’ of things, but in this case, the ‘why’ is very important,” Church said, explaining how this project is part of an ongoing effort to improve the safety, productivity, and flexibility of biotechnology.
“These results might also open a whole new chemical toolbox for biotech production,” said Isaacs. “For example, adding durable polymers to a therapeutic molecule could allow it to function longer in the human bloodstream.”
But to have such an impact, the researchers said, large swaths of the genome need to be changed all at once.
“If we make a few changes that make the microbe a little more resistant to a virus, the virus is going to compensate. It becomes a back and forth battle,” Church said. “But if we take the microbe offline and make a whole bunch of changes, when we bring it back and show it to the virus, the virus is going to say ‘I give up.’ No amount of diversity in any reasonable natural virus population is going to be enough to compensate for this wildly new genome.”
In the first study, with just a single codon removed, the genomically recoded organism showed increased resistance to viral infection. With several additional codons reassigned, a “wildly new genome” would make it impossible for engineered genes to escape into wild populations, Church said, because they would be incompatible with natural genomes. This could be of considerable benefit with strains engineered for drug or pesticide resistance, for example. What’s more, incorporating rare, non-standard amino acids could ensure strains only survive in a laboratory environment.
Engineering and evolution
Since a single genetic flaw can spell death for an organism, the challenge of managing a series of hundreds of specific changes was daunting, the researchers said. In both projects, the researchers paid particular attention to developing a methodical approach to planning and implementing changes and troubleshooting the results.
“We wanted to develop the ability to efficiently build the desired genome and to very quickly identify any problems — from design flaws or from undesired mutations — and develop workarounds,” Lajoie said.
The team relied on number of technologies developed in the Church lab and the Wyss Institute and with partners in academia and industry, including next-generation sequencing tools, DNA synthesis on a chip, and MAGE and CAGE genome editing methods. But one of the most important tools they used was the power of natural selection, the researchers added.
“When an engineering team designs a new cellphone, it’s a huge investment of time and money. They really want that cell phone to work,” Church said. “With E. coli we can make a few billion prototypes with many different genomes, and let the best strain win. That’s the awesome power of evolution.”
Funding was from Department of Energy, NSF, NIH, DARPA, Arnold and Mabel Beckman Foundation, Department of Defense NDSEG Fellowship, NSF graduate fellowship, NIH Director’s Early Independence Award, U.S. Office of Naval Research, Agilent Technologies, Wyss Institute, Department of Defense NDSEG Fellowship, and Air Force Contract #FA8721-05-C-0002.
Abstract of Science paper
We describe the construction and characterization of a genomically recoded organism (GRO). We replaced all known UAG stop codons in Escherichia coli MG1655 with synonymous UAA codons, which permitted the deletion of release factor 1 and reassignment of UAG translation function. This GRO exhibited improved properties for incorporation of nonstandard amino acids that expand the chemical diversity of proteins in vivo. The GRO also exhibited increased resistance to T7 bacteriophage, demonstrating that new genetic codes could enable increased viral resistance.
(¯`*• Global Source and/or more resources at http://goo.gl/zvSV7 │ www.Future-Observatory.blogspot.com and on LinkeIn Group's "Becoming Aware of the Futures" at http://goo.gl/8qKBbK │ @SciCzar │ Point of Contact: www.linkedin.com/in/AndresAgostini
 Washington
Washington