How Internet surveillance predicts disease outbreak before WHO
Creating your own disease surveillance maps
January 20, 2014
[+]
Have you ever Googled for an online diagnosis before visiting a
doctor? If so, you may have helped provide early warning of an
infectious disease epidemic.
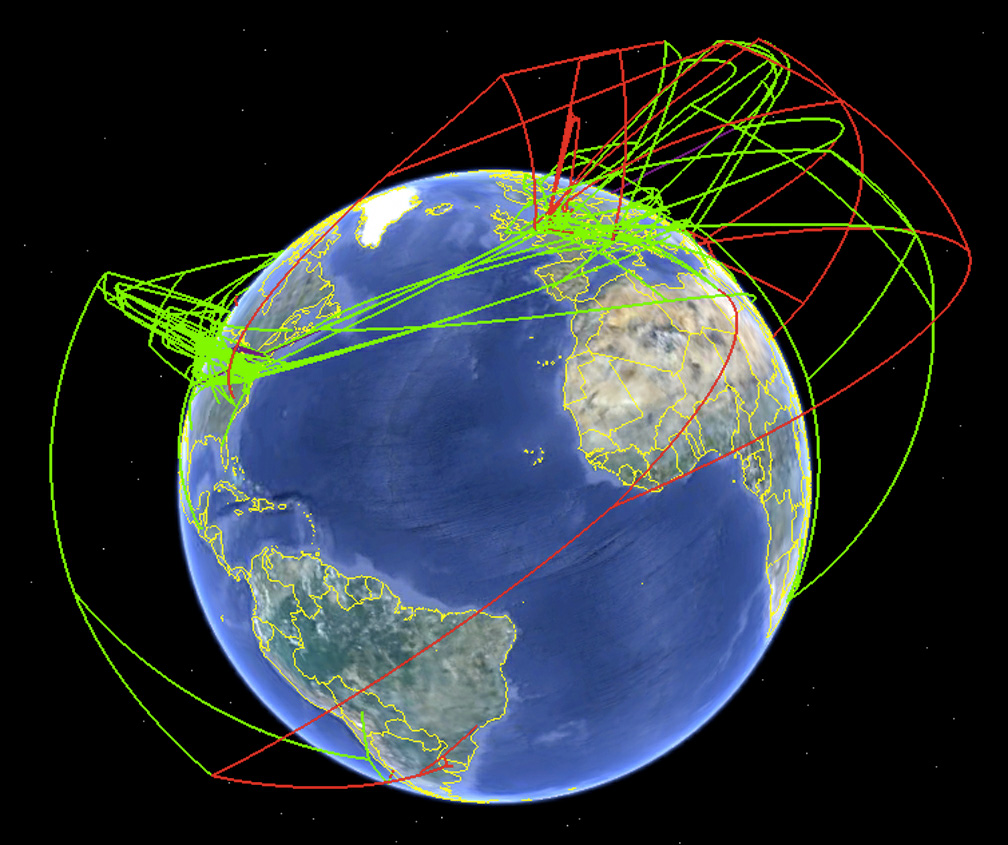
Screenshot
of the spread of H7 influenza as produced by Supramap and visualized by
Google Earth in 2009. This view illustrates the historical spread of
high pathogenic lineages (high-altitude red lines) and the local
evolution of high pathogenicity (low-altitude red lines). (Credit:
Janies/OSU)
In a new study published in Lancet Infectious Diseases, Internet-based surveillance has been found to detect infectious diseases such as Dengue Fever and Influenza up to two weeks earlier than traditional surveillance methods, according to Queensland University of Technology (QUT) research fellow and senior author of the paper Wenbiao Hu.
Hu, based at the Institute for Health and Biomedical Innovation, said there was often a lag time of two weeks before traditional surveillance methods could detect an emerging infectious disease.
“This is because traditional surveillance relies on the patient recognizing the symptoms and seeking treatment before diagnosis, along with the time taken for health professionals to alert authorities through their health networks. In contrast, digital surveillance can provide real-time detection of epidemics.”
Hu said the study used search engine algorithms such as Google Trends and Google Insights. It found that detecting the 2005–06 avian influenza outbreak “Bird Flu” would have been possible between one and two weeks earlier than official surveillance reports.
“In another example, a digital data collection network was found to be able to detect the SARS outbreak more than two months before the first publications by the World Health Organization (WHO),” Hu said.
[+]
“Early detection means early warning and that can help reduce or
contain an epidemic, as well alert public health authorities to ensure
risk management strategies such as the provision of adequate medication
are implemented.”
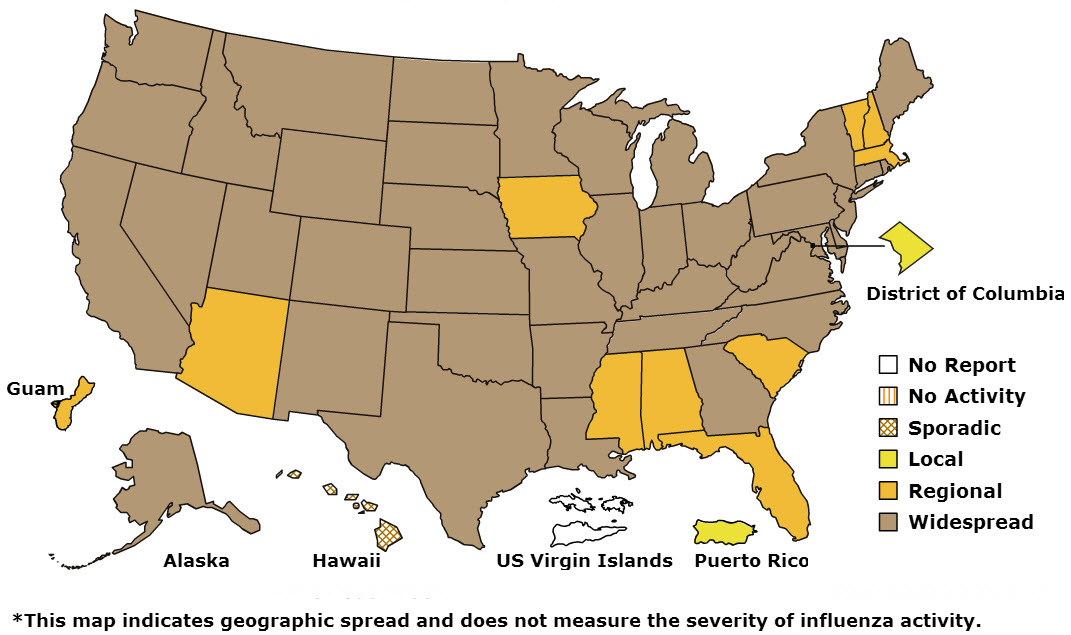
According
to this week’s CDC FluView report published Jan. 17, 2014, influenza
activity in the United States remains high overall, with 3,745
laboratory-confirmed influenza-associated hospitalizations reported since October 1, 2013 (credit: CDC)
Hu said the study found that social media including Twitter and Facebook and microblogs could also be effective in detecting disease outbreaks. “The next step would be to combine the approaches currently available such as social media, aggregator websites, and search engines, along with other factors such as climate and temperature, and develop a real-time infectious disease predictor.”
“The international nature of emerging infectious diseases combined with the globalization of travel and trade, have increased the interconnectedness of all countries and that means detecting, monitoring and controlling these diseases is a global concern.”
The other authors of the paper were Gabriel Milinovich (first author), Gail Williams and Archie Clements from the University of Queensland School of Population, Health and State.
Supramap
Another powerful tool is Supramap, a web application that synthesizes large, diverse datasets so that researchers can better understand the spread of infectious diseases across hosts and geography by integrating genetic, evolutionary, geospatial, and temporal data. It is now open-source — create your own maps here.
Tracking Swine Flu: Daniel Janies, former
associate professor of Biomedical Informatics at The Ohio State
University, shows how he used Google Earth to track the H1N1 flu virus.
(2009)
It was originally developed in 2007 to track the spread and evolution of pandemic (H1N1) and avian influenza (H5N1).
“Using SUPRAMAP, we initially developed maps that illustrated the spread of drug-resistant influenza and host shifts in H1N1 and H5N1 influenza and in coronaviruses, such as SARS,” said Janies. “SUPRAMAP allows the user to track strains carrying key mutations in a geospatial browser such as Google Earth. Our software allows public health scientists to update and view maps on the evolution and spread of pathogens.”
Grant funding through the U.S. Army Research Laboratory and Office supports this Innovation Group on Global Infectious Disease Research project. Support for the computational requirements of the project comes from the American Museum of Natural History (AMNH) and OSC. Ohio State’s Wexner Medical Center, Department of Biomedical Informatics and offices of Academic Affairs and Research provide additional support.
Abstract of The Lancet Infectious Diseases paper
Emerging infectious diseases present a complex challenge to public health officials and governments; these challenges have been compounded by rapidly shifting patterns of human behaviour and globalisation. The increase in emerging infectious diseases has led to calls for new technologies and approaches for detection, tracking, reporting, and response. Internet-based surveillance systems offer a novel and developing means of monitoring conditions of public health concern, including emerging infectious diseases. We review studies that have exploited internet use and search trends to monitor two such diseases: influenza and dengue. Internet-based surveillance systems have good congruence with traditional surveillance approaches. Additionally, internet-based approaches are logistically and economically appealing. However, they do not have the capacity to replace traditional surveillance systems; they should not be viewed as an alternative, but rather an extension. Future research should focus on using data generated through internet-based surveillance and response systems to bolster the capacity of traditional surveillance systems for emerging infectious diseases.
Abstract of The Supramap project: linking pathogen genomes with geography to fight emergent infectious diseases
Novel pathogens have the potential to become critical issues of national security, public health and economic welfare. As demonstrated by the response to Severe Acute Respiratory Syndrome (SARS) and influenza, genomic sequencing has become an important method for diagnosing agents of infectious disease. Despite the value of genomic sequences in characterizing novel pathogens, raw data on their own do not provide the information needed by public health officials and researchers. One must integrate knowledge of the genomes of pathogens with host biology and geography to understand the etiology of epidemics. To these ends, we have created an application called Supramap (http://supramap.osu.edu) to put information on the spread of pathogens and key mutations across time, space and various hosts into a geographic information system (GIS). To build this application, we created a web service for integrated sequence alignment and phylogenetic analysis as well as methods to describe the tree, mutations, and host shifts in Keyhole Markup Language (KML). We apply the application to 239 sequences of the polymerase basic 2 (PB2) gene of recent isolates of avian influenza (H5N1). We map a mutation, glutamic acid to lysine at position 627 in the PB2 protein (E627K), in H5N1 influenza that allows for increased replication of the virus in mammals. We use a statistical test to support the hypothesis of a correlation of E627K mutations with avian-mammalian host shifts but reject the hypothesis that lineages with E627K are moving westward. Data, instructions for use, and visualizations are included as supplemental materials at: http://supramap.osu.edu/sm/supramap/publications.
(¯`*• Global Source and/or more resources at http://goo.gl/zvSV7 │ www.Future-Observatory.blogspot.com and on LinkeIn Group's "Becoming Aware of the Futures" at http://goo.gl/8qKBbK │ @SciCzar │ Point of Contact: www.linkedin.com/in/AndresAgostini
 Washington
Washington